Long COVID’s Viral Persistence Hypothesis: Scientific Sticking Points
TLDR: The viral persistence theory of long COVID could benefit greatly from better standardization of detection methods both in tissue and blood. We have solidly identified a few sites of viral persistence in long COVID patients but the full range is still unclear. Characterizing tissue-specific viral persistence can provide relevant information at this point. While animal and human cell models can help narrow our search and dissect mechanistic details, it’s still difficult to interpret findings with our current level of knowledge. I include tables of viral persistence models and of criteria I look for when evaluating viral persistence models.

I’ve been working on projects related to viral persistence and these thoughts stem from following online discourse and challenges I’ve directly encountered.
First off, what is long COVID’s viral persistence theory? The viral persistence theory suggests that the lingering symptoms experienced by some people after recovering from the acute phase of COVID-19 are due to the continued presence of SARS-CoV-2, or parts of it, in the body. It centers on this idea that virus, or maybe just fragments of its genetic material, remains in certain cells or tissues, leading to potentially ongoing inflammation or alterations in important metabolites.
There have been big bets made hinging on this theory. There are multiple clinical trials for antivirals such as Paxlovid aiming to target these hypothetical persistent reservoirs in long COVID patients. Given the importance of this theory to current long COVID research, what are some of the challenges facing viral persistence research?
Sticking Points Searching For Reservoirs
Detecting virus in autopsies, biopsies, and surgeries from the general population has shed light on potential reservoirs of SARS-CoV-2 in humans. However, identifying the virus in living long COVID patients presents challenges with a limited number of tissue types attainable.1 Patient samples obviously inform our interpretation and development in vivo and in vitro models of viral persistence. So what barriers exist?
Obtaining tissues from biobank repositories with pertinent clinical information is its own logistic challenge. Institutional Review Board (IRB) protocols detailing your search for virus need to address potential viral exposure. Safety protocols are needed when dealing with tissue from severe COVID-19 cases but what about when it’s uncertain if virus is in tissue? This isn’t a huge deal if all your samples are fixed with something like formalin as this should inactivate the virus but that in turn limits the type of analysis that can be done (particularly RNAseq/scRNAseq). Being overly cautious greatly limits the amount of samples which can be collected while being under-cautious potentially exposes investigators to harm.
Searching For A Needle In A Haystack
Obviously, using a technique that maximizes sensitivity/specificity and amount of tissue tested while minimizing cost is ideal. Yet, it isn’t clear what that technique is at the moment. Investigators use a wide variety of techniques to detect viral RNA or protein (mainly RNA sequencing, PCR, immunohistochemistry, fluorescence in situ hybridization, nCounter, and electron microscopy2). Results from these tests aren’t synonymous–detecting viral RNA vs viral protein vs competent virus leads to different interpretations. Furthermore, ambiguous detection at the analytic limits of the technique muddies the waters and has led to suggestions that persistent virus is not necessarily highly replicating.
Standardizing guidelines and performing head-to-head comparisons for these various methods has a host of benefits—reducing false positives, improving reliability of results, and clarifying some of the ambiguity investigators face. Having specific antibodies, primers, or FISH probes which have been heavily vetted takes out a lot of the guesswork. These three studies here, here, and here address a few of these issues:
.jpg)
In line with this, establishing secondary validation methods to verify positive results is part of this due diligence. Ultimately, having field-wide best practices lowers barriers to entry for labs who may have access to such incidental tissues. For our proverbial haystack, having more searchers also increases your odds of finding a needle.
Results will likely not be as conclusive as you would hope. If you don’t find virus in these samples, you’ll question the sensitivity of your methods, whether you didn’t test enough of your sample, or if the reservoir is in a part of the tissue you didn’t sample. If you do detect virus, heterogeneity introduced by tissue sampling and long COVID sub-types will complicate interpretation when some patients or controls are positive and others are negative (a probable scenario).
Conflicting evidence from other investigators will similarly fuel speculation. Do some patients have different or absent reservoirs? Evidence suggests otherwise healthy people can have viral persistence—why don’t they have long COVID? Case reports or series reporting viral detection in long COVID autopsies/biopsies/surgeries are a starting point that will provide plausibility but will suffer in terms of generalizability.
These issues really highlight the need for comprehensive, non-invasive evaluation of persistent virus. Emerging detection methods funded by PolyBio, such as bio-sensor T cells and non-invasive scans, will become extremely important for the identification of patient reservoirs if they work on the scale we need them to work. These semi-recent results show promise.
Sticking Points Modeling SARS-CoV-2 Persistence
Many labs driving our knowledge of long COVID biology approach it through the lens of immunology or pathology.3 This work has been the key to understanding empiric features of long COVID so far. However, it feels like these approaches are reaching their limits. While these studies are undoubtedly valuable, more studies focused on the same readily measurable blood/plasma immunological parameters won’t move the needle of our understanding like the past ones did.
Up to this point, virologists have been more peripherally involved, characterizing infection models but unsure of whether or not findings relate to long COVID. There are two approaches to evaluate potential mechanisms of long COVID:
Infecting models and correlating post acute findings with human data (e.g. infecting a mouse and seeing if it develops long COVID like symptoms)
Simulating hypothesized pathologic causes and observing the effect in models (e.g. trying to establish persistent gut infection and seeing what changes this causes4)
For the latter, there are a few hypothesized long COVID mechanisms that are amenable to basic virology research–chiefly herpesvirus reactivation and viral persistence. The viral persistence theory is the more straightforward gateway for virology labs into this field and I predict we’ll start seeing more publications from these labs in the near future. Notably, this has been done for other human coronaviruses like 229E and OC43 can persist in a wide variety of culture and animal models. Honestly, the absence of analogous models for SARS-CoV-2 is somewhat perplexing given the preponderance of evidence supporting viral persistence in humans.
Both animal and human cell models are important, each addressing different aspects of the “where” and “why” in the context of viral persistence. While human cells better address “where?”, in vivo animal models are better suited for assessing “why?” despite intrinsic anatomic/immune/viral receptor differences with humans. You can establish more representative infectious foci with human cell models but will lack the ability to link traveling disease mediators and complex symptoms together like you can with animals. Ideally, we would get data on a tissue’s capacity for viral persistence from both of these systems, using patient samples to resolve conflicting results.
Persistent models naturally take more time–at odds with the rapid rate of viral evolution. This poses challenges as variants or sublineages come and go in the time it takes to thoroughly develop the project. Deciding which variants to focus on is a non-trivial aspect with each included variant greatly increasing the amount of experiments involved. We have some idea that different variants pose different risks of long COVID but we haven’t been able to narrow down this effect to specific viral features (another facet virologists are suited to study).
Persistent Cell Cultures
| Cell Type | Duration | Study |
|---|---|---|
| Human airway epithelial cells (air-liquid interface) with multiple genotypes | 51 days | Link |
| In vitro differentiated human nasal epithelial cells and bronchial epithelial cells | 28 days | Link |
| MucilAirTM primary human bronchial epithelial cells | 21 days | Link (Supplementary Figure 7) |
| iPSC-derived human midbrain organoids | 28 days | Link |
| C2BBe1 enterocyte cell line | 23 days | Link |
| iPSC-derived human cardiac microtissues | 28 days | Link |
| Vero E6 (adaptation study) | 42 days (probably most cells died ~7 days in and the surviving cells re-expanded) | Link |
Like patient samples, human-derived models like cell lines, ex vivo tissue, and organoids have been important in establishing the potential tropism of SARS-CoV-2.5 Such studies can be easily adapted to evaluate viral persistence.
Currently, what are the best candidates to test for persistence? In no particular order: lymphatic tissue, intestines, respiratory tract, heart, vasculature, and brain. These tissues have all demonstrated varying degrees of infectability and persistence in autopsy/biopsy samples. For the lungs, heart, and intestines, these midline structures all receive and transmit information through the vagus nerve, a key player that could link tissue-level infection, immunity, and the central nervous system.
How long is persistent? Can we even measure persistence in certain models? Generally, cells (especially differentiated or primary cells) have time frames in which they are viable and maintain their appropriate phenotype, barring the use of finicky systems that die too quickly or de-differentiate. Additionally, most of the common cell lines (A549, VeroE6) used for viral characterization demonstrate massive cell death with cytolysis and synctia formation somewhere between 5 and 14 days post-infection. The above table shows the longest cultures I’ve encountered in the literature, all around ~30 days (which in my mind is still squarely within the time frame of a normal acute infection). In some studies I’m working on, I settled on 60 days as the final evaluation point–hitting the two month mark for long COVID symptom duration in the WHO definition while being manageable experimentally.
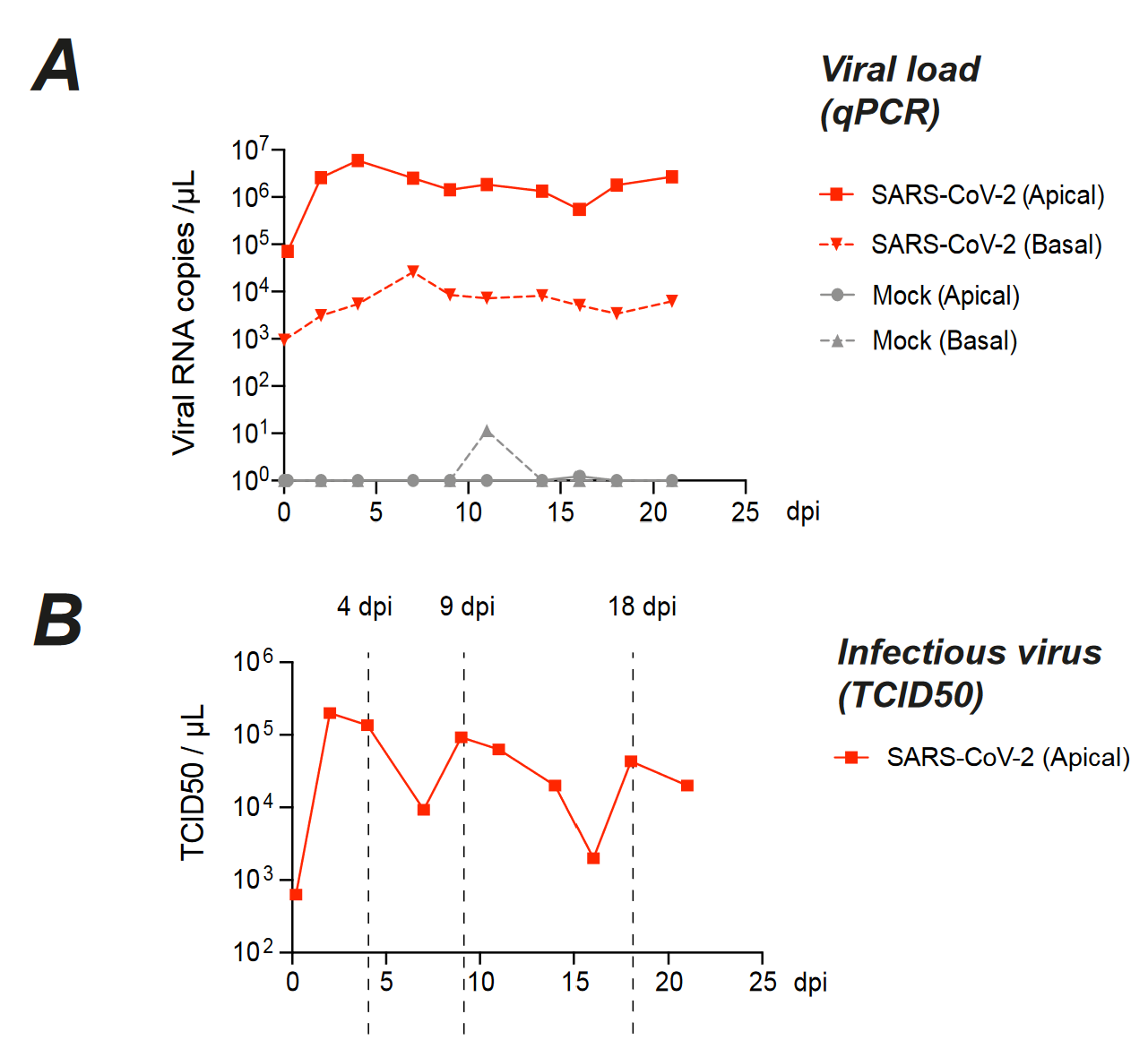
Other criticisms. If possible, testing persistence in multiple cell models of the same tissue can alleviate criticism that results are cell line specific or genetic background specific. Unless you seed your culture or organoid with immune cells, their absence in most culture models means persistent virus seen in your model may be cleaned up early on in real patients. The argument that persistence may be an artifact of the cell culture system is difficult to counter without associated evidence of persistence in patients.
Persistent Animal Models
| Animal | Tissue | Duration | Study |
|---|---|---|---|
| Macaques | Lungs | 18 months | Link |
| Rhesus macaques and African green monkeys | Brain | Up to 27-29 days | Link |
| Humanized MISTRG6 mice with lung-specific hACE2 | Lungs | At least 35 days | Link |
| Humanized DRAGA mice expressing hACE2 | No direct data shown | At least 25 days | Link |
| K18-hACE2 mice with Calu-3 xenografts | Human lung tumor cells | 30 days | Link |
| CD4+/CD8+ T cell-depleted C57BL/6J mice infected with Beta variant (has N501Y mutation) | Nose | At least 28 days | Link |
| C57BL/6J mice infected with mouse-adapted SARS-CoV-2 MA10 | Lung, Nose | Mostly non-detectable past ~5-7 days | Link |
| BALB/c mice infected with mouse-adapted SARS-CoV-2 MA10 | Lung, Nose | Mostly non-detectable past ~5-15 days | Link, Link |
| Syrian golden hamster | Lung, nose, trachea, brian | Minimally detectable by 14 days | Link |
Difficulties with translation. Long COVID trials are happening before the development of solid, pre-clinical animal models. Many of the symptoms (particularly the cognitive ones) are difficult to accurately assess in animals. As clinically-focused teams are sorting out human biomarkers, it is unclear if the biomarkers detected in long COVID patient cohorts translate to animal models (I’m sure someone is working on this with the huge, recent improvements in this area). Surveying the many animal species for signs of long COVID-like disease presentations could also lead us to a good albeit unconventional model.
Do you ever think about the animals who may have ME/CFS & Long Covid?
— emily fraser 🌿🐌✨ (@emilyesfraser) January 4, 2024
Viral spread throughout the body is naturally impacted by the amount of virus used to inoculate animals. In humans, long COVID can happen after both severe and mild infections, now in individuals with some degree of prior immunity or cross-immunity. Most models are going to use animals without such prior immunity to SARS-CoV-2 or other coronaviruses. Using too much virus will kill most of them while using too little may prevent them from developing long-term sequelae. So what viral titer do we use? With either outcome, it is difficult to get an adequate sample size without using large numbers of animals.
If we conclusively knew the nature and site of viral persistence in patients, we could simply replicate these findings in mice by resolving viral tropism and immune differences. This entails confirming the virus has the possibility of infecting the same sites as in humans and that viral control by the immune system (or lack of control) is similar. In the absence of clear human data, researchers did not initially consider tissue-level viral persistence to be a feature of disease worth designing models around. I think that is changing given the consistency of human findings. For example, designing a gut specific viral persistence model could address questions about long COVID, viral evolution (those cryptic lineages that keep showing up in wastewater surveillance), and transmissibility of gut-derived virus.
There is a very delicate balance that must be struck here. It’s not simply about having virus persist but about having virus persist in an otherwise immunocompetent model. Impairing facets of innate and adaptive immunity IS likely to create chronically infected animal models; however, these models are more likely to resemble persistently detectable, chronic infections in immunocompromised patients rather than the sneaky, relatively covert viral persistence implicated in long COVID.
Notes on rodent models. Based on the literature I’ve seen, humanization of mice (beyond expression of human ACE2) seems to lead to increased viral persistence. Granted, many studies aren’t evaluating the same time points but this was a general trend I saw looking through papers. On the other hand, mice adapted MA10 virus models (adapting the virus to the animal) are being used to study post-viral symptoms but consistently demonstrate clearance of virus after roughly a week. Similarly, Syrian golden hamster models which are otherwise excellent models of acute COVID pathogenicity clear virus in similar time frames.
If we can get the tropism and dynamics of infection right in rodents, there are so many neat tricks that can be employed to dissect long COVID disease mechanisms. For example, non-invasive imaging of reporter viruses in living animals could be used to localize and track viral persistence over long time frames as long as the reporter gene remains stably expressed. Animal models potentially allow differentiation between direct effects of a persistent viral reservoir and secondary effects, such as immune dysregulation. For rodent models, humanization of the immune system or tissue-selective human ACE2 expression could further refine models of viral persistence. Hot take: the SARS-CoV-2 field in general needs to move away from the K18-hACE2 mouse model towards models that express human ACE2 at sites and levels similar to humans.
I predict more and more animal models of “long COVID” will publish results this year, the majority of which will be in K18-hACE2 mice and Syrian golden hamsters (what’s most widely available). I suspect most if not all of these models will demonstrate complete clearance of virus in the early post-acute stages. While I believe these models have merit in understanding subtypes of long COVID without viral persistence, there will undeniably be a discordance between the direction human studies are pointing us (viral persistence) and what will be investigated by these studies.
What I Expect From A Truly Persistent Cell Culture / Animal Model
| Experimental Factor | Proposed Standard |
|---|---|
| Length | Greater than or equal to 30 days but ideally greater than or equal to 60 days. |
| Amount of Virus | 0.01< Multiplicity of Infection < 1 for cell lines/organoids (MOI 5 is pretty high to me already); Viral titer will vary for different animal models. I think both titers that cause mild and severe disease are valid for animal models. |
| Variants Tested | One variant is fine if it’s interesting. Otherwise, at least the original and a semi-recent Omicron sublineage like BA.5, XBB, etc. (later than BA.1 - BA.2 is fine). |
| Viral Detection | Evaluate viral RNA, protein, and competent virus. Ideally detection of nucleocapsid. Subgenomic RNA optional but would be nice. |
| Passaging | Applies to cultures only. No passaging onto fresh, un-infected cells - only splitting if needed. |
| Animal Model | Open to pretty much any model but minus points if it’s in K18-hACE2 mice. |
Organizing these considerations, here are criteria I’ll be using to evaluate viral persistence models. I’m sure there will be great, reliable viral persistence studies that don’t meet all of these criteria. However, I’ll be pining for what could have been, longing for a version of the study that addressed all of these points.
Wrap Up
If you’re someone who’s studying SARS-CoV-2 viral persistence–what’s your experience been like? What do you think of these criteria? What are sticking points that can be improved in conducting this research?

MD/PhD Student at University of Texas Medical Branch
I write about viruses, data science, and ballet.
Click home at the top of the page to see other things I’ve been working on.
Footnotes
This PolyBio project is looking to collect lymph node and gut samples from long COVID patients.↩︎
See the controversy about prior studies erroneously claiming normal organelles were virus particles here↩︎
There are obvious exceptions like Dr. Akiko Iwasaki’s lab that integrates immunology, pathology, and virology.↩︎
See the recent serotonin paper with a nuanced discussion by Dr. Janna Moen↩︎
In some cases identifying infectable sites before detection in patients. Signs of choroid plexus infection in humans was found after organoids demonstrated potential susceptibility.↩︎