Currently: MD-PhD Student...
Keywords: Infectious Disease, Pathology, SARS-CoV-2, Flaviviruses, Organoids, Viral CNS Infections, Viral Recombination, Bioinformatics, Data Science, Ballet, Equity in the Arts, AAPI Creative Pipeline
Former Ballet Dancer → Future Physician Scientist
My biomedical research is centered on viral diseases. While I no longer dance professionally, I coordinate research focused on understanding disparities and long-term outcomes in ballet.
I combine skill sets in virology 🦠, medicine ⚕️, and data science 📈. I am currently doing my PhD in Dr. Pei-Yong Shi’s and Dr. Xuping Xie’s lab where I work with SARS-CoV-2.
Interests
Most of my current work uses reverse genetics systems and in vitro models to characterize cloned virus mutants. The Dr. Pei-Yong Shi lab uses this approach to study drug resistance in support of novel therapies, viral evolution in the context of emerging variants, and how protein structure relates to function.
Xia, H.*, Yeung, J.*, Kalveram, B.*, Bills, C.J., Chen, J.Y.-C., Kurhade, C., Zou, J., Widen, S.G., Mann, B.R., Kondor, R., Todd Davis, C., Zhou, B., Wentworth, D.E., Xie, X., Shi, P.Y., 2023. Cross-neutralization and viral fitness of SARS-CoV-2 Omicron sublineages. Emerging Microbes & Infections 0, 1–19. Link
Yeung, J., Wang, T., Shi, P.-Y., 2023. Improvement of mucosal immunity by a live-attenuated SARS-CoV-2 nasal vaccine. Current Opinion in Virology 62, 101347. Link
Adam, A., Kalveram, B., Chen, J.Y.-C., Yeung, J., Rodriguez, L., Singh, A., Shi, P.-Y., Xie, X., Wang, T., 2023. A single-dose of intranasal vaccination with a live-attenuated SARS-CoV-2 vaccine candidate promotes protective mucosal and systemic immunity. npj Vaccines 8, 1–6. Link
Bills, C.J., Xia, H., Chen, J.Y.-C., Yeung, J., Kalveram, B., Walker, D., Xie, X., Shi, P.-Y., 2023. Mutations in SARS-CoV-2 variant nsp6 enhance type-I interferon antagonism. Emerging Microbes & Infections 0, 2209208. Link
*Contributed Equally
Recombination is an essential driver of virus evolution and adaption, giving rise to new chimeric viruses, structural variants, sub-genomic RNAs, and Defective-RNAs. Next-Generation Sequencing of virus samples, either from experimental or clinical settings, has revealed a complex distribution of recombination events that contribute to the intrahost diversity within individual hosts. This diversity has been associated with viral persistence, immune evasion, and termination of infection in different contexts.
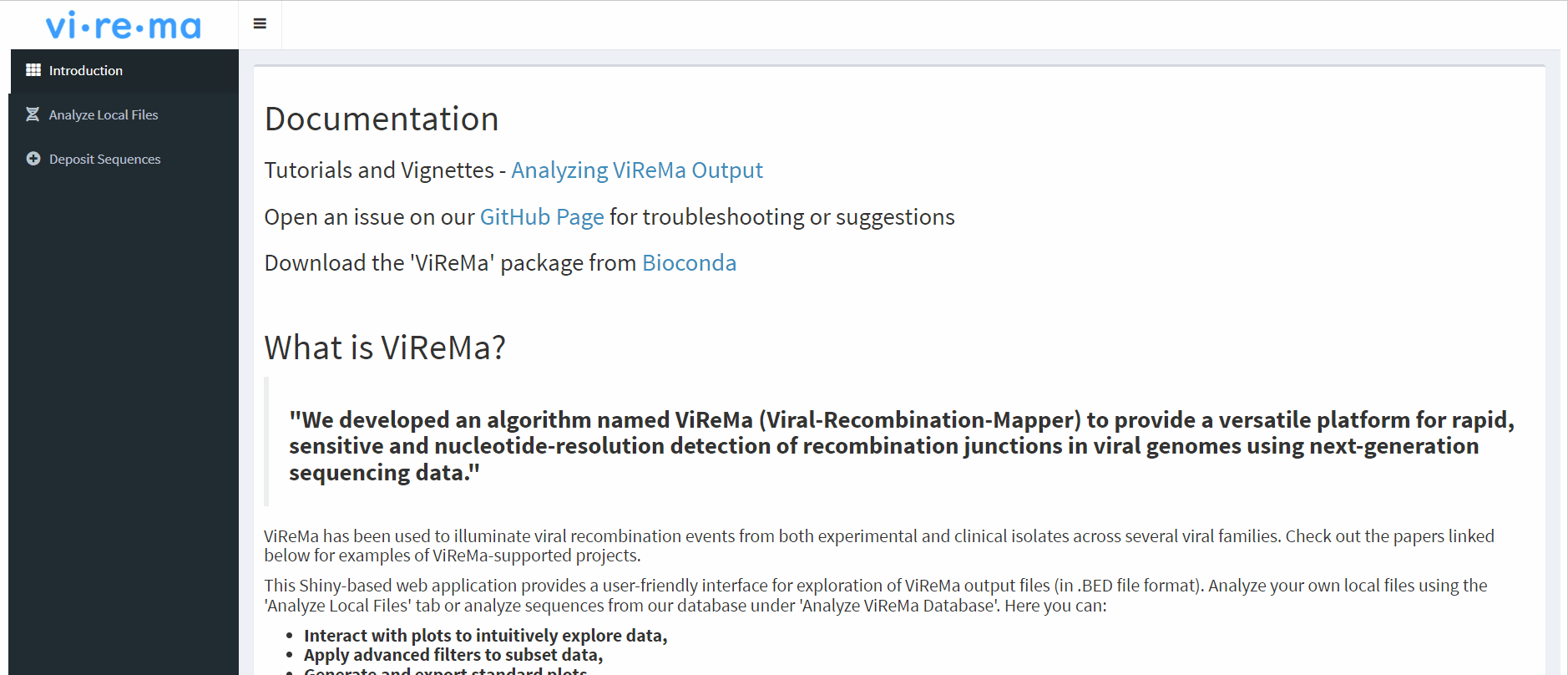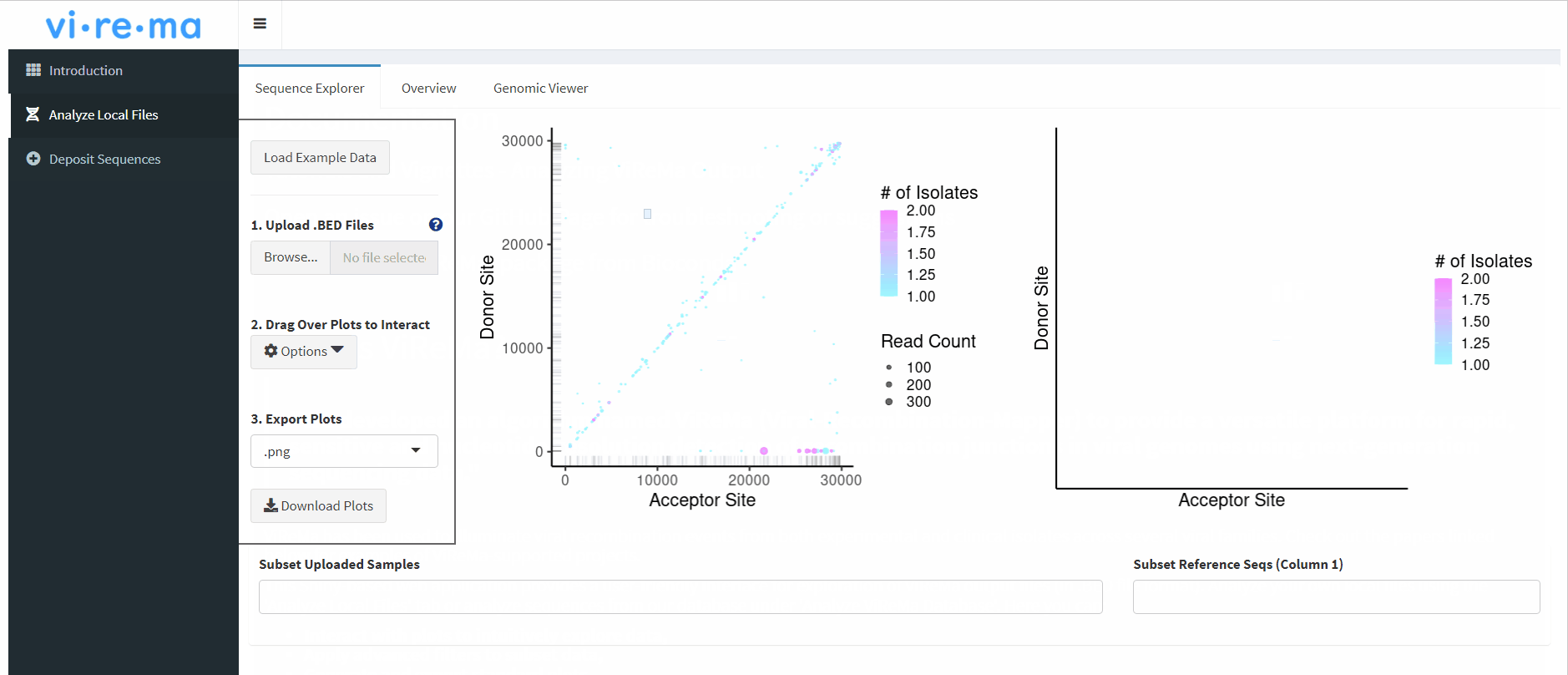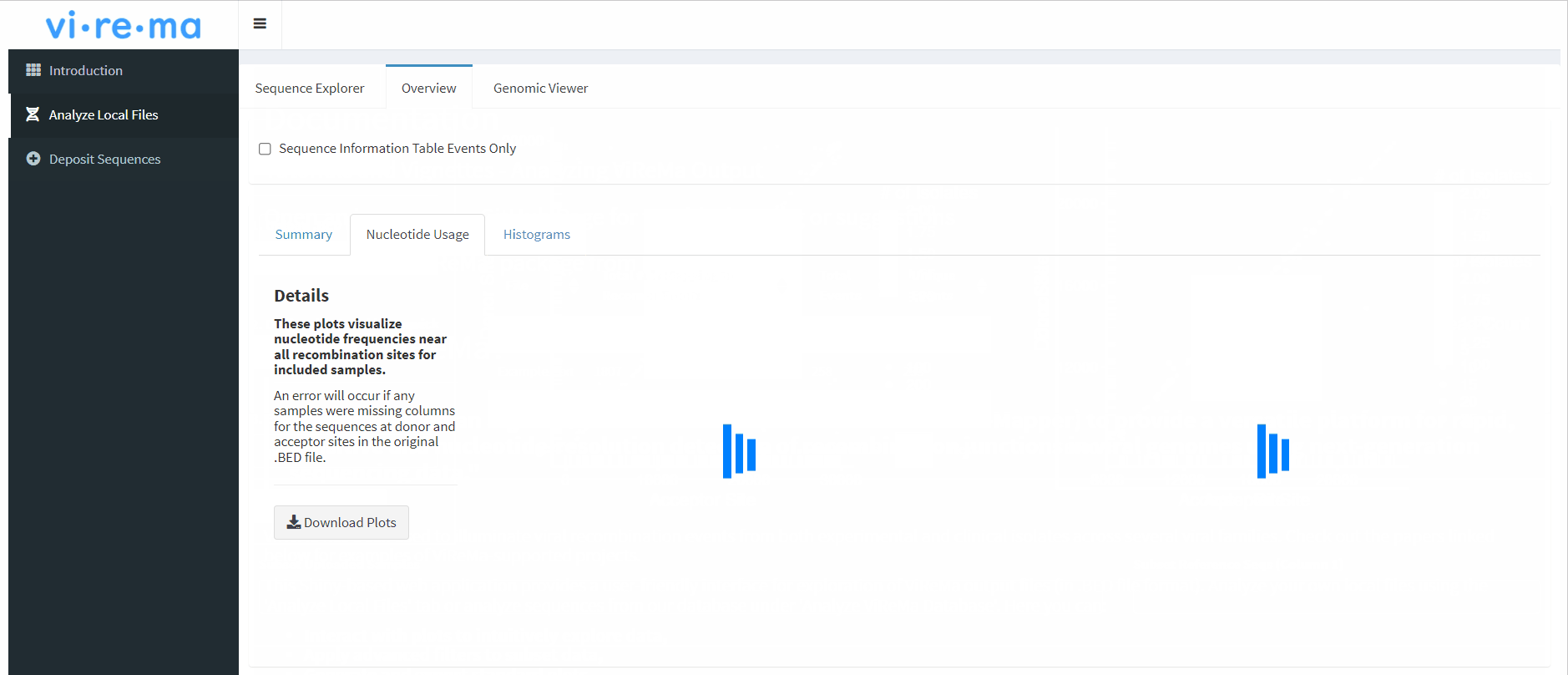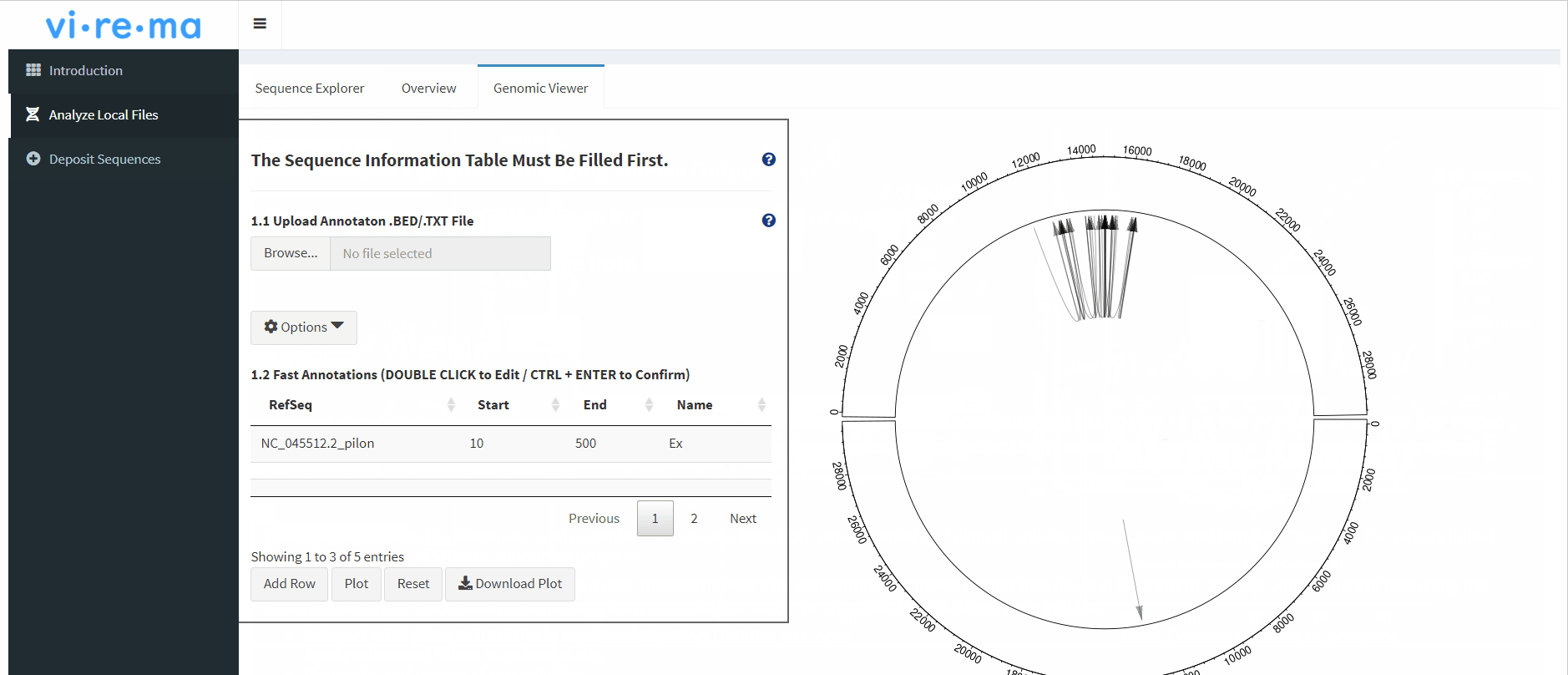
I have worked with Dr. Andrew Routh to standardize and expand the detection of non-homologous recombination in viral sequencing data. So far, this has involved making a point-and-click analysis tool in R Shiny and containerized versions of the detection software ViReMa. We are working on new workflows for detecting viral structural variants and applying them to publicly available data sets on the Sequence Read Archive. This will enhance the discovery of such events, allow correlation with disease-relevant outcomes, and enable exploration of intrinsic properties of recombination across virus families, conditions, and time points.
Yeung J, Routh AL: ViReMaShiny: an interactive application for analysis of viral recombination data. Bioinformatics 2022, 38:4420–4422. Link
Sotcheff S, Zhou Y, Yeung J, Sun Y, Johnson JE, Torbett BE, Routh AL: ViReMa: a virus recombination mapper of next-generation sequencing data characterizes diverse recombinant viral nucleic acids. GigaScience 2023, 12, giad009. Link
Viral infections of the central nervous system (CNS) have a wide spectrum of clinical severity. While some viruses can cause profound architectural and behavioral changes, others are asymptomatic or cause subtle cognitive deficits.
Modeling infections of human brains is greatly potentiated by organoid technology. My work with the Dr. Ernesto Marques and Vishwajit Nimgaonkar labs used cortical organoids to investigate HSV-1 and Zika virus infection. For Zika virus, we explored changes in the complement system upon CNS infection as complement has an important role in neurodegeneration, neuro-development, and synaptic pruning. In order to recapitulate complement protein production in the brain, we used a non-organoid culture model to inform the development of a pluripotent stem cell derived model that would more accurately reflect expression profiles seen in humans. We also studied changes in complement production and surface-bound modulators after Zika infection.
With Dr. Pei-Yong Shi’s lab, I am currently investigating the impact of SARS-CoV-2 infection on brain organoids and potential connections to Long COVID. We are also collaborating with various partners to study viral persistence in CNS tissue reservoirs.
Zheng W, Klammer AM, Naciri JN, Yeung J, Demers M, Milosevic J, Kinchington PR, Bloom DC, Nimgaonkar VL, D’Aiuto L: Patterns of Herpes Simplex Virus 1 Infection in Neural Progenitor Cells. Journal of Virology 2020, 94:e00994-20. Link
Marques ETA, Demers M, D’Aiuto L, Castanha PMS, Yeung J, Wood JA, Chowdari KV, Zheng W, Yolken RH, Nimgaonkar VL: Herpesvirus Infections in the Human Brain: A Neural Cell Model of the Complement System Derived from Induced Pluripotent Stem Cells, in: Current Topics in Behavioral Neurosciences. Springer, Berlin, Heidelberg, pp. 1–22. Link
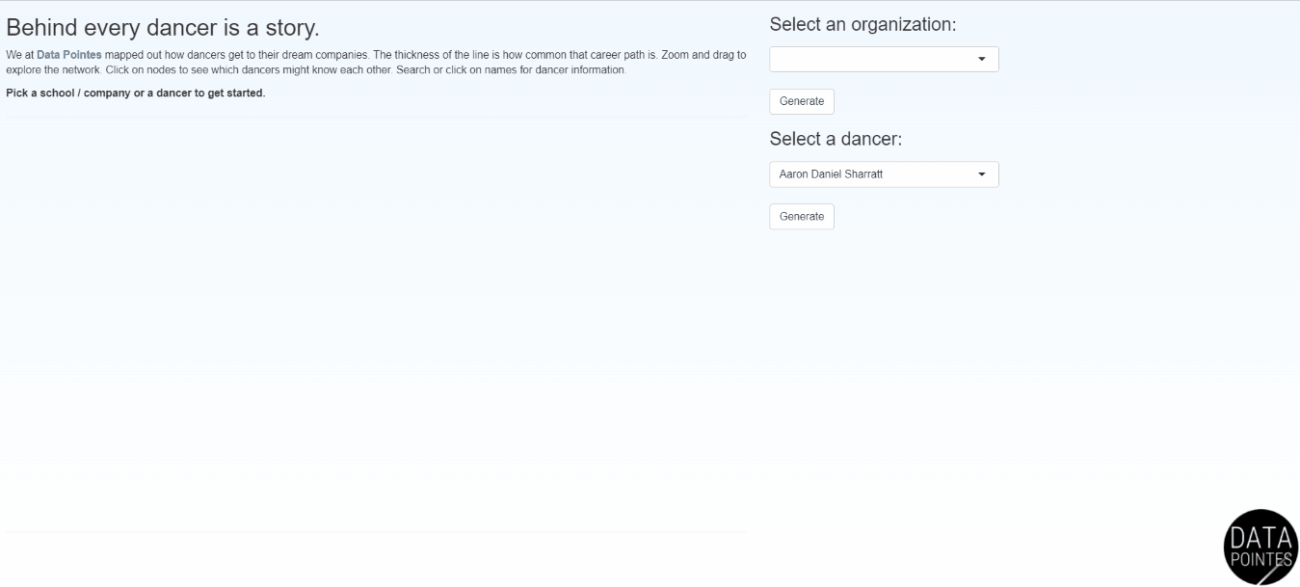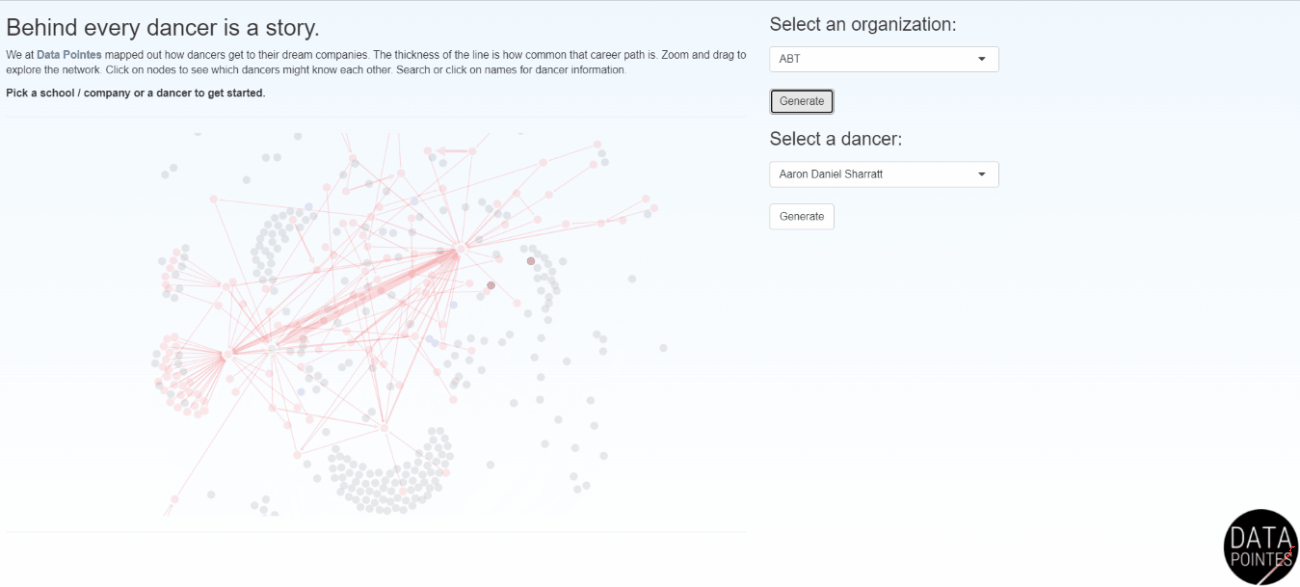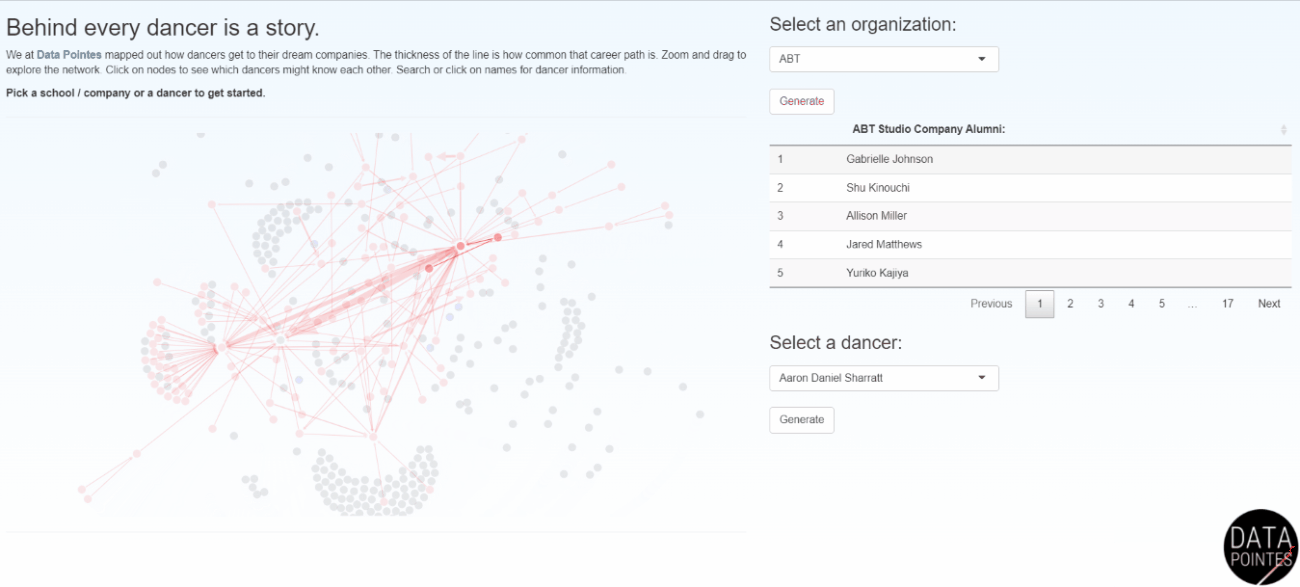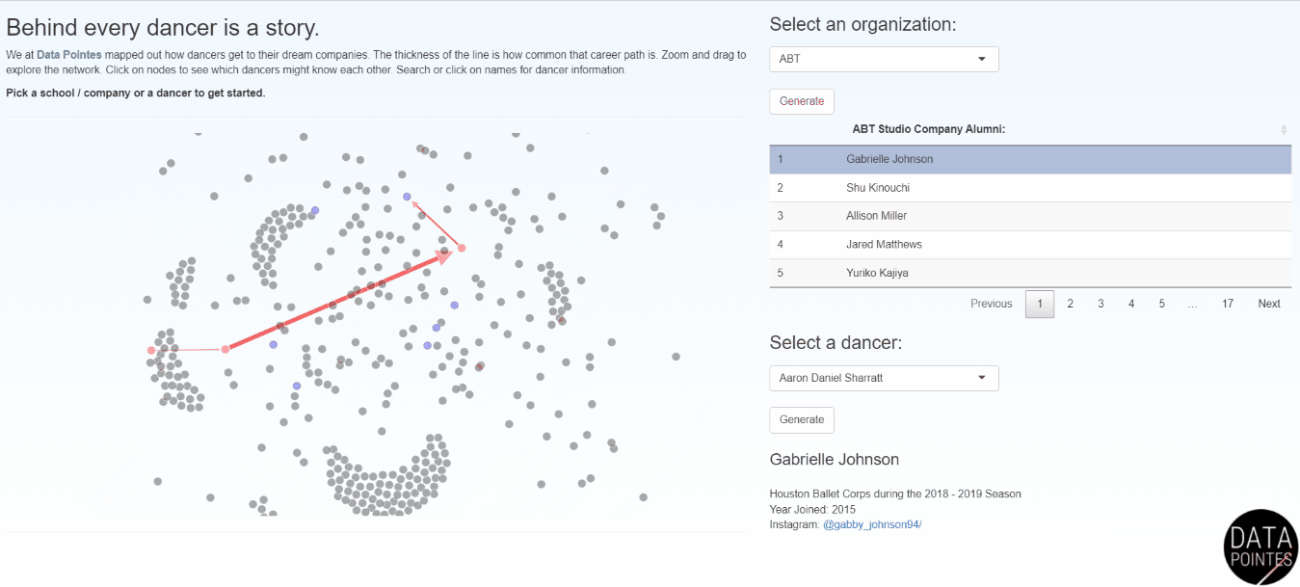
Becoming a professional ballet dancer is an extremely competitive endeavor. Navigating this path is difficult without insider knowledge and there is little transparency among training organizations. I use data science skills to provide insights into the long-term outcomes of ballet dancers. Data Pointes centers around the analysis of multiple datasets I maintain and the website gets thousands of unique visits a year. Dozens of parents have said Data Pointes provides a starting point for objective discussions about their child’s path to professional ballet.
I work with non-profit and academic partners that are interested in disparities in American ballet. Dance Data Project focuses on the underpromotion of women to leadership, lack of protective policies, and wage disparities in these ballet companies (I currently serve on the board). Gold Standard Arts Foundation is aiming to understand the Asian American and Pacific Islander (AAPI) ballet pipeline and launched the Final Bow for Yellowface initiative which helped modernize depictions of Asian cultures onstage.
Listen to me talk about why/how I started Data Pointes and the relevance of findings to the ballet world on the Ballet Help Desk Podcast:
Portfolio
Education
University of Texas Medical Branch | MD-PhD | 2020 -
✅ University of Pittsburgh | MS | 2018 - 2020
✅ Texas A&M University | BS | 2015 - 2018
Experience
Research using wet and dry lab techniques with experience at Biosafety Level 3. Over 5+ years building with R. Experience with D3.js, Python, Docker, Linux, and MongoDB. Currently learning Rust 🦀!